(sub-U04153.aparc.native.dlabel.nii)
(sub-U04153.aparc.native.dlabel.nii)
For context, two T1w 0.8mm isotropic MPRAGE images were collected per subject. This was done to achieve the higher 0.8mm resolution (which has a poor signal-to-noise ratio), and allow for averaging out the noise by combining the images afterward.
The documentation for fMRIPrep claims it will average all found T1w images into a single reference template. This would be exactly what we want to happen.
But I have been skeptical that this actually happened in real life because of the recon-all command issued.
See below for full details.
In the output logs from fMRIPrep 20.2.0, run on the MRI Unit cluster as a docker service,
searching for the two T1w filenames yields:
406: N4BiasFieldCorrection -d 3 \
--input-image /input/sub-U04153/ses-20352/anat/sub-U04153_ses-20352_run-01_T1w.nii.gz \
--output sub-U04153_ses-20352_run-01_T1w_corrected.nii.gz
411: N4BiasFieldCorrection -d 3 \
--input-image /input/sub-U04153/ses-20352/anat/sub-U04153_ses-20352_run-02_T1w.nii.gz \
--output sub-U04153_ses-20352_run-02_T1w_corrected.nii.gz
414: lta_convert --inlta identity.nofile \
--outlta /tmp/work/fmriprep_wf/single_subject_U04153_wf/anat_preproc_wf/anat_template_wf/t1w_conform_xfm/mapflow/_t1w_conform_xfm1/out.lta \
--src /input/sub-U04153/ses-20352/anat/sub-U04153_ses-20352_run-02_T1w.nii.gz \
--trg /input/sub-U04153/ses-20352/anat/sub-U04153_ses-20352_run-02_T1w.nii.gz
419: lta_convert --inlta identity.nofile \
--outlta /tmp/work/fmriprep_wf/single_subject_U04153_wf/anat_preproc_wf/anat_template_wf/t1w_conform_xfm/mapflow/_t1w_conform_xfm0/out.lta \
--src /input/sub-U04153/ses-20352/anat/sub-U04153_ses-20352_run-01_T1w.nii.gz \
--trg /input/sub-U04153/ses-20352/anat/sub-U04153_ses-20352_run-01_T1w.nii.gz
Searching for the N4 corrected filename yields:
497: mri_robust_template --satit --fixtp \
--mov /tmp/work/fmriprep_wf/single_subject_U04153_wf/anat_preproc_wf/anat_template_wf/n4_correct/mapflow/_n4_correct0/sub-U04153_ses-20352_run-01_T1w_corrected.nii.gz \
/tmp/work/fmriprep_wf/single_subject_U04153_wf/anat_preproc_wf/anat_template_wf/n4_correct/mapflow/_n4_correct1/sub-U04153_ses-20352_run-02_T1w_corrected.nii.gz \
--inittp 1 --iscale --noit --template sub-U04153_ses-20352_run-01_T1w_template.nii.gz --subsample 200 \
--lta /tmp/work/fmriprep_wf/single_subject_U04153_wf/anat_preproc_wf/anat_template_wf/t1w_merge/tp1.lta \
/tmp/work/fmriprep_wf/single_subject_U04153_wf/anat_preproc_wf/anat_template_wf/t1w_merge/tp2.lta
This is clear evidence that fMRIPrep created a combined image,
called sub-U04153_ses-20352_run-01_T1w_template.nii.gz.
So I then searched for that combined file, skipped several lines to get to my original source of skepticism...
3187: recon-all -autorecon1 \
-i /tmp/work/fmriprep_wf/single_subject_U04153_wf/anat_preproc_wf/anat_template_wf/t1w_merge/sub-U04153_ses-20352_run-01_T1w_template.nii.gz \
-T2 /input/sub-U04153/ses-20352/anat/sub-U04153_ses-20352_T2w.nii.gz \
-noskullstrip -hires -openmp 7 -subjid sub-U04153 -sd /output/freesurfer \
-expert /tmp/work/fmriprep_wf/single_subject_U04153_wf/anat_preproc_wf/surface_recon_wf/autorecon1/expert.opts
So my skepticism was based on the fact recon-all was fed a file named ...run-01...,
but from the lines above, it is clear that the ...run-01... file is the mean of both runs,
but was poorly named by fMRIPrep.
So fMRIPrep has co-registered and averaged the two T1w images in this case. Great! But when I looked at the file fed to FreeSurfer from the fMRIPrep pipeline and from the ABCD pipeline, ABCD is much better. So it appears fMRIPrep may have done the right thing, but done it poorly.
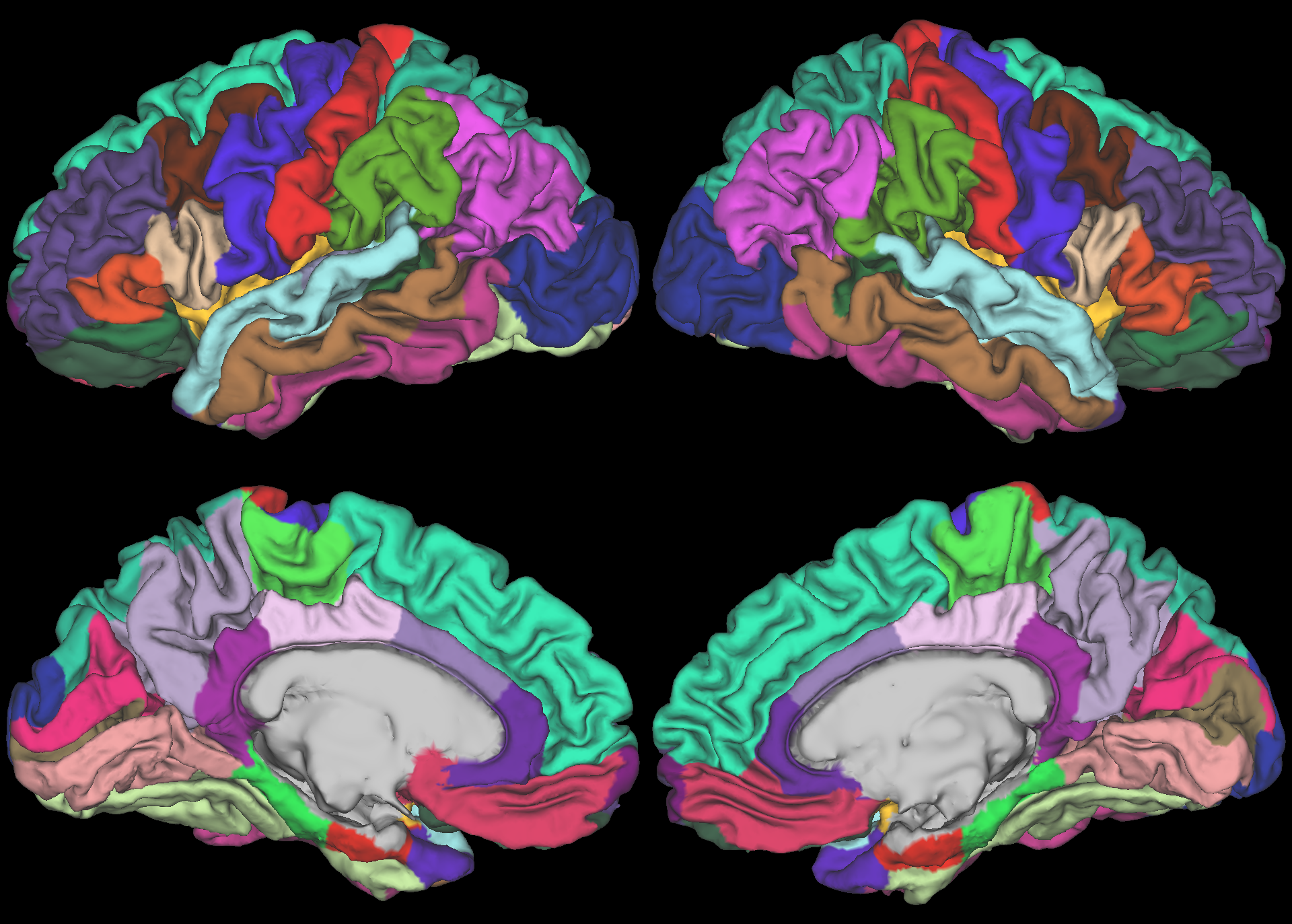
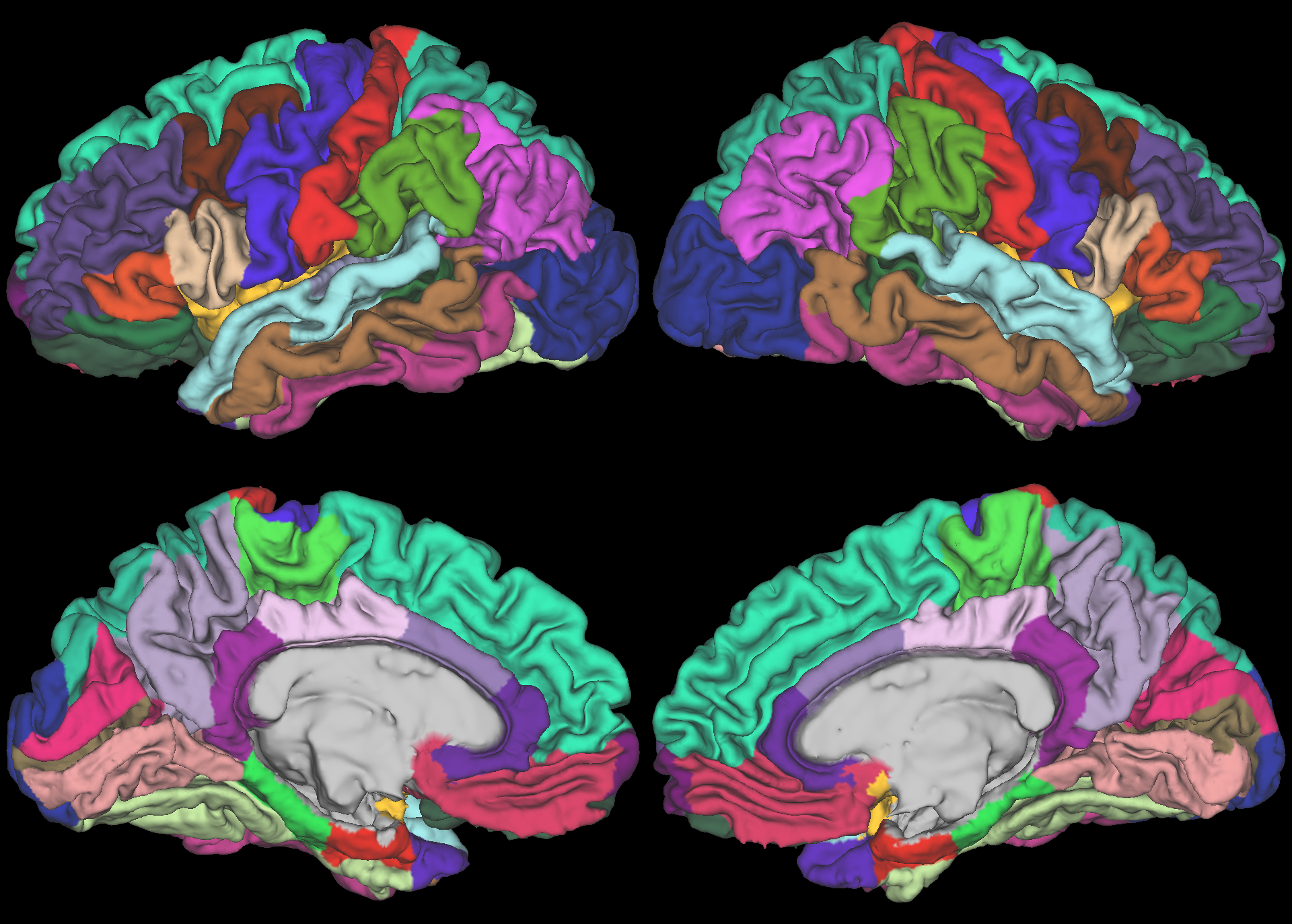
Parcellations are very similar, with some possible exception between insula wrapping under to medial orbitofrontal cortex.
| ABCD Cortical Parcellation (sub-U04153.aparc.native.dlabel.nii) |
Ciftify, post-fMRIPrep, Cortical Parcellation (sub-U04153.aparc.native.dlabel.nii) |
|---|---|

|

|